Section for Microbial and Chemical Ecology

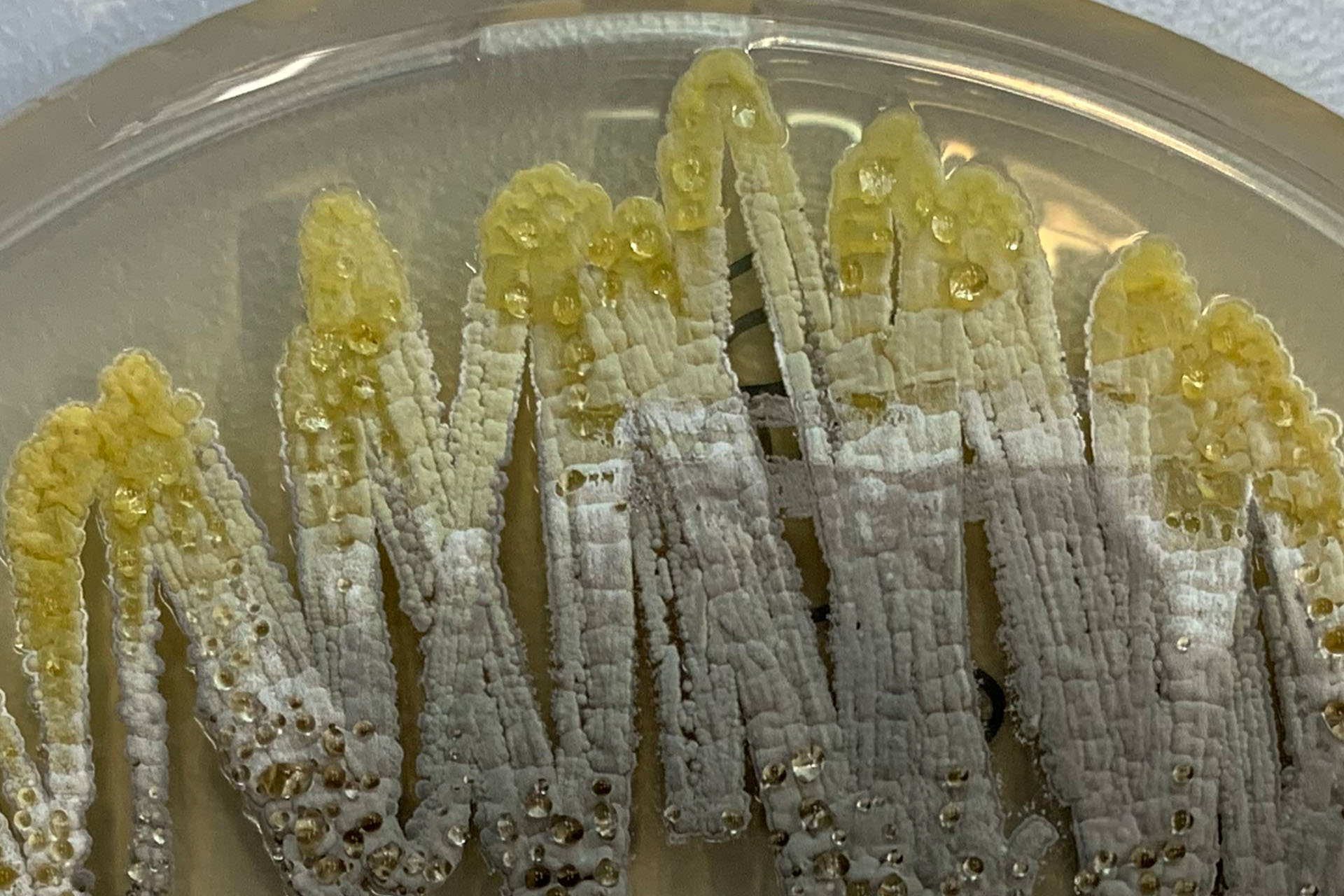
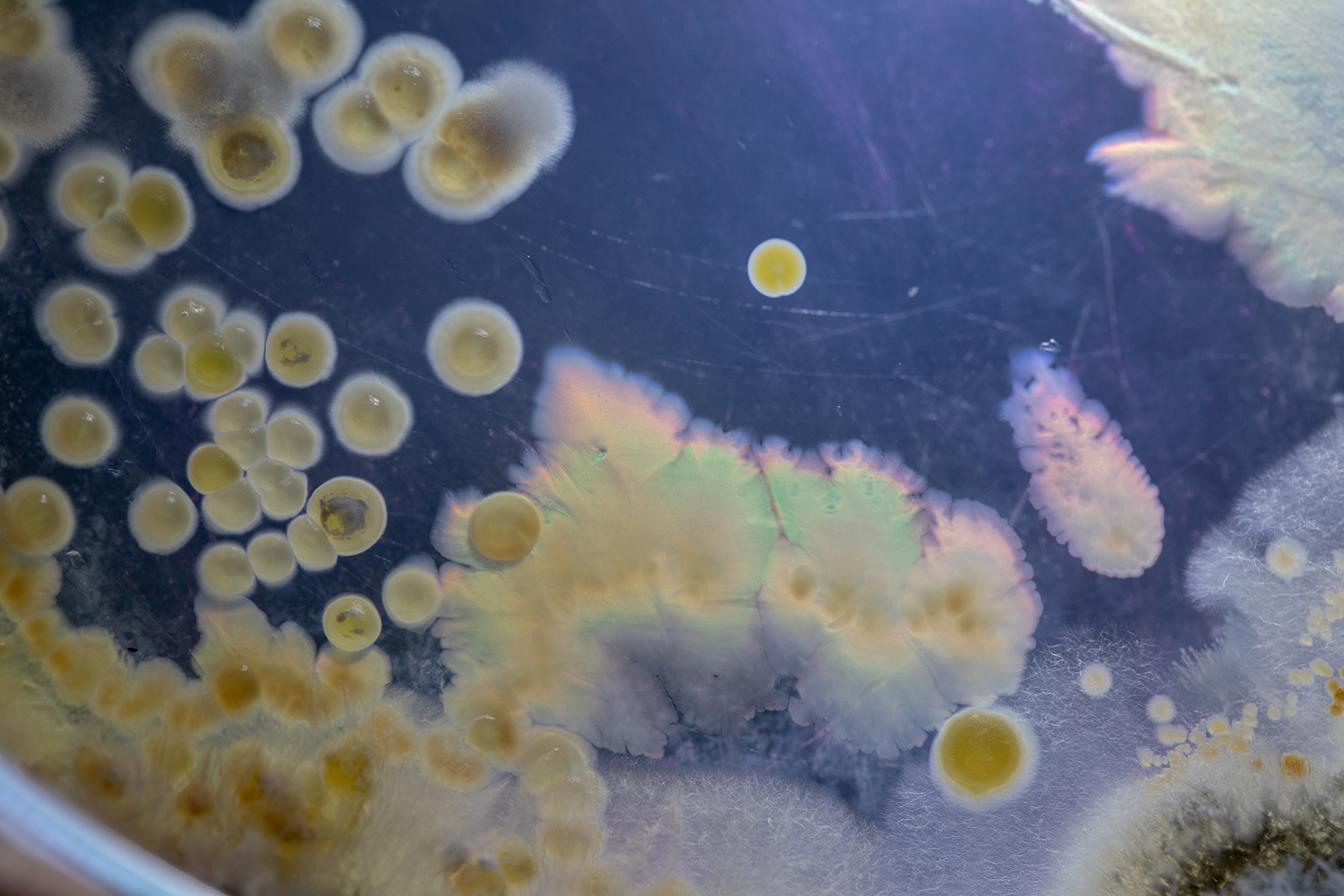
Microorganisms occupy almost every niche available on this planet and serve important functions of both benefit and challenge to mankind. In almost all niches, microorganisms live in mixed, complex communities, which, when found in association with higher organisms have been described as “microbiomes”.
Understanding and engineering both pure cultures and microbiomes in natural settings and in production is a key focus of the section. Collectively, this will enable us to discover and design new strategies, bioactive compounds or biologicals that can be used to e.g. combat diseases in plants, animals and humans, design new ways of degrading unwanted compounds in nature, develop novel strains in biotechnology or use novel substrates in the biomanufacturing process.
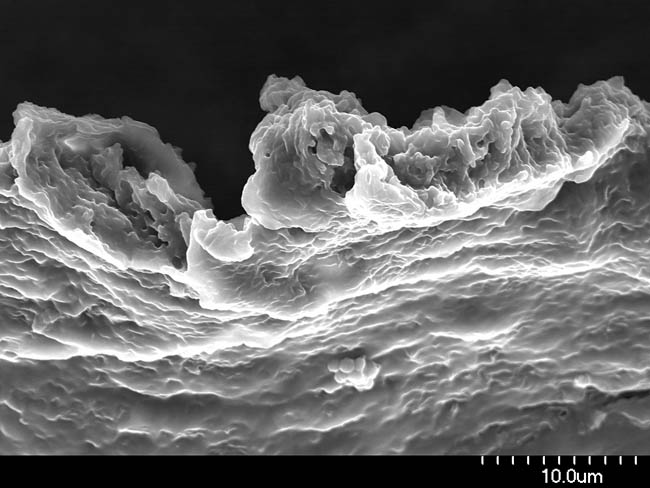
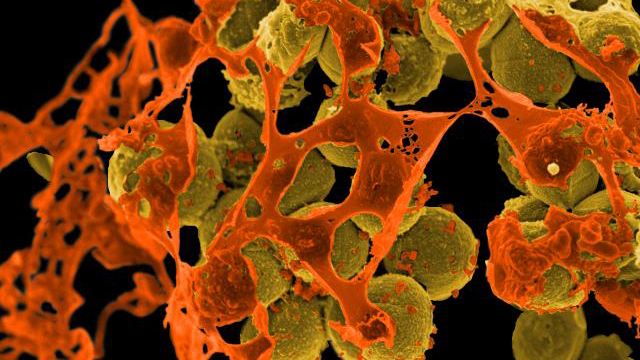
The MCE section studies and uses pure cultures of microorganisms and microbiomes with desirable properties such as probiotic activity, production of bioactive secondary metabolites, pigments or degradative enzymes. In addition, the section applies basic insight into natural microbial habitats and exploit physiological, metabolic and functional diversity to combat pathogenic bacteria and filamentous fungi and their toxin production. Microbial diversity, evolution and community dynamics are studied by both classical microbiological, genetic and biochemical approaches, advanced molecular profiling, genomics, transcriptomics, proteomics, metabolomics, and bioinformatics as well as functional and structural characterization of purified key molecules.
Understanding and engineering both pure cultures and microbiomes in natural settings and in production
Head of Section
Lone Gram Professor Mobile: +45 23688295 gram@bio.dtu.dk