Enzyme Discovery
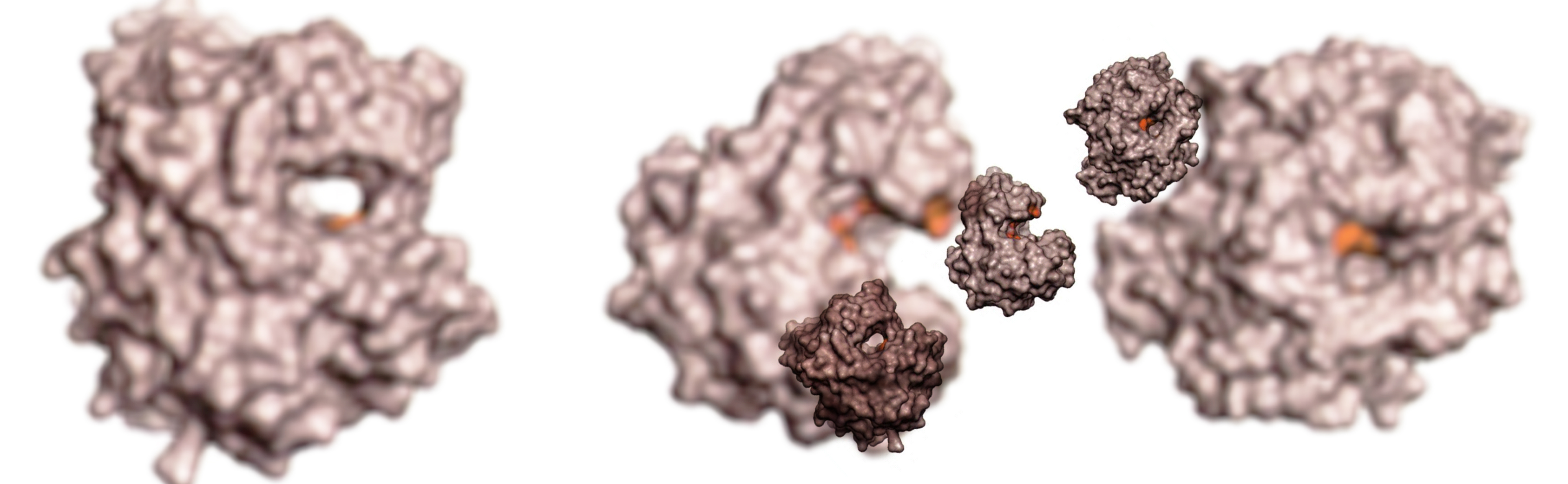
Assigning precise function to genomic data is one of the greatest scientific challenges of modern Biology. Genomes encode a large diversity of enzymes with high substrate specificities such as carbohydrate-active enzymes (CAZymes). The exceptional diversity of their substrates makes CAZymes powerful tools to explore omics data and explain a range of biological processes.
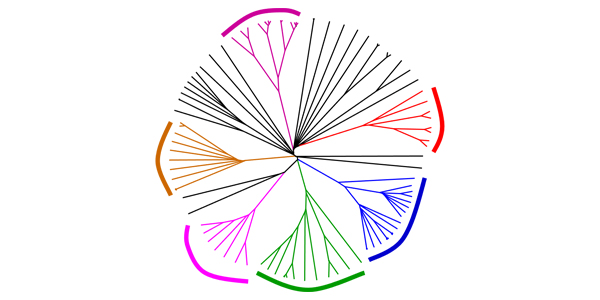
Sequence-based categorization groups CAZymes together in families that have related three-dimensional folds and catalytic mechanisms, but members of the same family frequently act on different substrates. The evolutionary processes by which CAZymes acquired novel specificities from older ones have left traces in the amino acid sequence that can be detected and exploited to ultimately predict the function of contemporary sequences.
However, the major bottlenecks for high accuracy prediction of the fine specificity of CAZymes from sequence data only are the incomplete knowledge of the extent of their substrates, the heterogeneous sampling of the sequence-space by experimentally characterized enzymes and the lack of suitable representations of the details of their specificity and mechanism for deep learning algorithms.
Novel enzymes and predictions from sequence data
We develop a synergistic, multidisciplinary program that utilizes high-throughput production and characterization of CAZymes to rationally explore the CAZyme sequence space not only to discover novel enzymes and enzyme families, but also to ultimately be able to accurately predict fine specificity from sequence data only. Our work finds applications in various areas, from the functional exploration of microbiomes to the identification of novel enzymes for biotech applications and biorefineries.