Marie Sofie Møller Junior Research Group
My research focusses on applied molecular enzyme chemistry. This involves studying protein-carbohydrate interactions with the aim of obtaining novel insights, which allow for obtaining enzymes with altered desired functions through engineering.
A number of different approaches and methods are used for pursuing this line of research:
- Heterologous gene expression in E. coli or Pichia pastoris
- Enzymatic activity analysis using reducing-end assays, thin layer chromatography and HPLC
- Deciphering the function of specific amino acid residues or protein domains through point mutations or truncations selected using bioinformatics and structure analysis as well as computational design using Rosetta
- Molecular interaction analysis using surface plasmon resonance (SPR), affinity gel electrophoresis and pull-down assays
- Protein stability analysis using e.g. differential scanning calorimetry
- Protein structure determination using X-ray crystallography
Current research projects
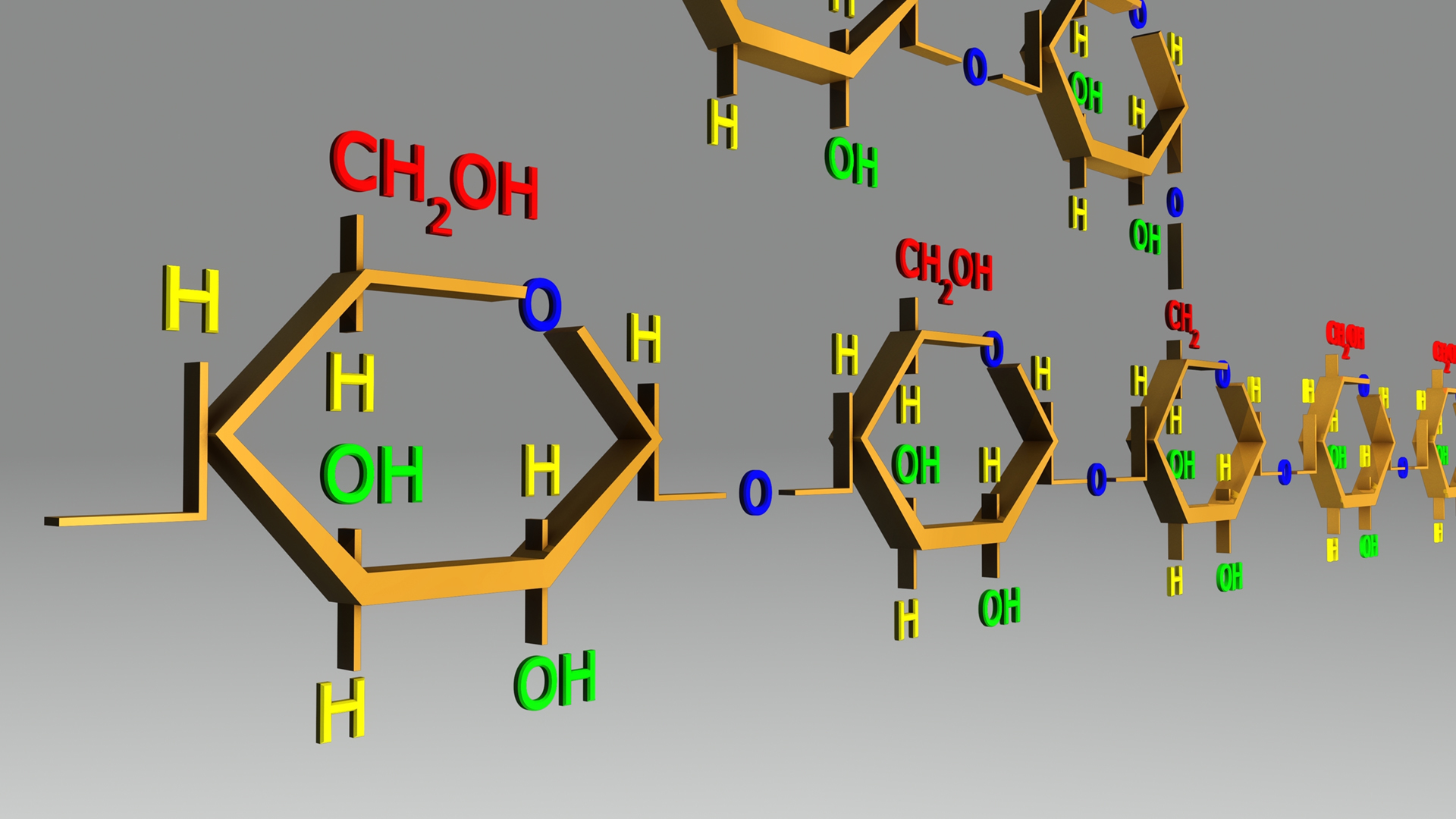
α-glucan debranching enzymes as tools for producing an array of valuable bioproducts
(Funded by the Novo Nordisk Foundation)
In nature and industry starch is degraded by a cascade of enzymes. Starch is composed of two types of polysaccharides made from glucose: the essentially linear amylose, and the branched amylopectin having maltooligosaccharide chains interconnected through branch points. Debranching enzymes (DBEs) remove the branches from amylopectin by hydrolysis. Remarkably, some DBEs can catalyse the reverse hydrolysis reaction and create branch points in so-called transglycosylation reactions. The present project is going to utilize such transglycosylation activity of DBEs to obtain starch-derived mostly small to medium sized products with novel structures and functions, e.g. slowly digested carbohydrates with health promoting effects and glycoengineered natural compounds (secondary metabolites) for bioplastic incorporation acting as natural pH indicators. A range of DBEs will be engineered to optimize transglycosylation specificity and yield, and to reduce (ideally abolishing) the hydrolytic reaction.
Contact
Marie Sofie Kjer Møller Associate Professor Department of Biotechnology and Biomedicine Phone: +45 45252741 msmo@dtu.dk
(Funded by Independent Research Fund Denmark, Technology and Production Sciences)
Man-made polymer materials like plastic are causing severe environmental problems. Enzymes capable of degrading plastic have been discovered, however, they are far from being efficient in industrial settings. One reason for this is that it is difficult for the enzymes to stick to the plastic. One solution is to exploit protein modules (carbohydrate-binding modules, CBMs), which nature uses for facilitating interactions between enzymes and recalcitrant polysaccharides (cellulose, starch, and chitin). A few CBMs have been shown to bind to plastics, but this binding promiscuity has not yet been studied systematically. Indeed CBMs have to be engineered in order to bind efficiently to different plastic types. Bioinformatics methods are here used for selecting at least 100 of these protein modules, which will be i) screened and ii) optimised for polymer binding ability. Both classical plastic types and bio-based composite plastics will be included. Some of the bio-based plastics available today can only be partially degraded in nature. Optimized protein modules will be fused with relevant polymer degrading enzymes. The aim is to obtain more efficient enzymes, which can solve the problem of sustainable waste management of man-made polymers.
