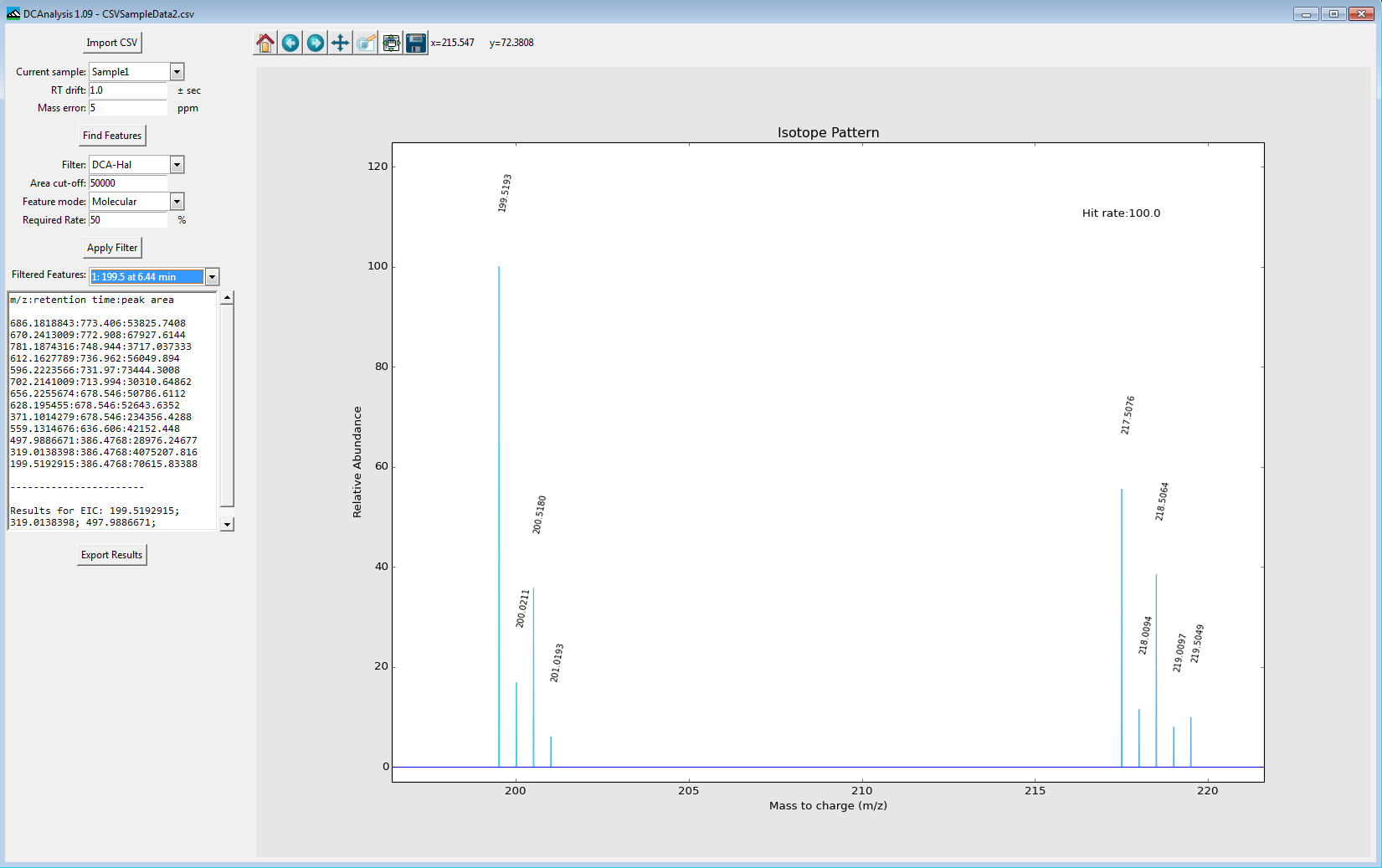
LC-HRMS Filtering: Dynamic Cluster Analysis
The DTU Metabolomics Core has developed dynamic cluster analysis (DCA), a series of filtering algorithms which can screen high resolution mass spectromic data for sulphur containing compounds (with DCA-Sul) and chlorine/bromine containing compounds (with DCA-Hal).
These algorithms have been incorporated into a programme call DCAnalysis, and it can be downloaded here, along with instructions on its use and test data. The algorithms are modelled using the Marinlit natural products database and have been tested on reference standards and algae extracts published in Analytical Chemistry. The filtering technique utilises the novel A+1 to A+2 isotope cluster spacing to distinguish A+2 element containing compounds from others compounds.
Also available here is IsoPat, an isotope pattern calculator that can be used to simulate coeluting compounds and to calculate isotopic enrichment.
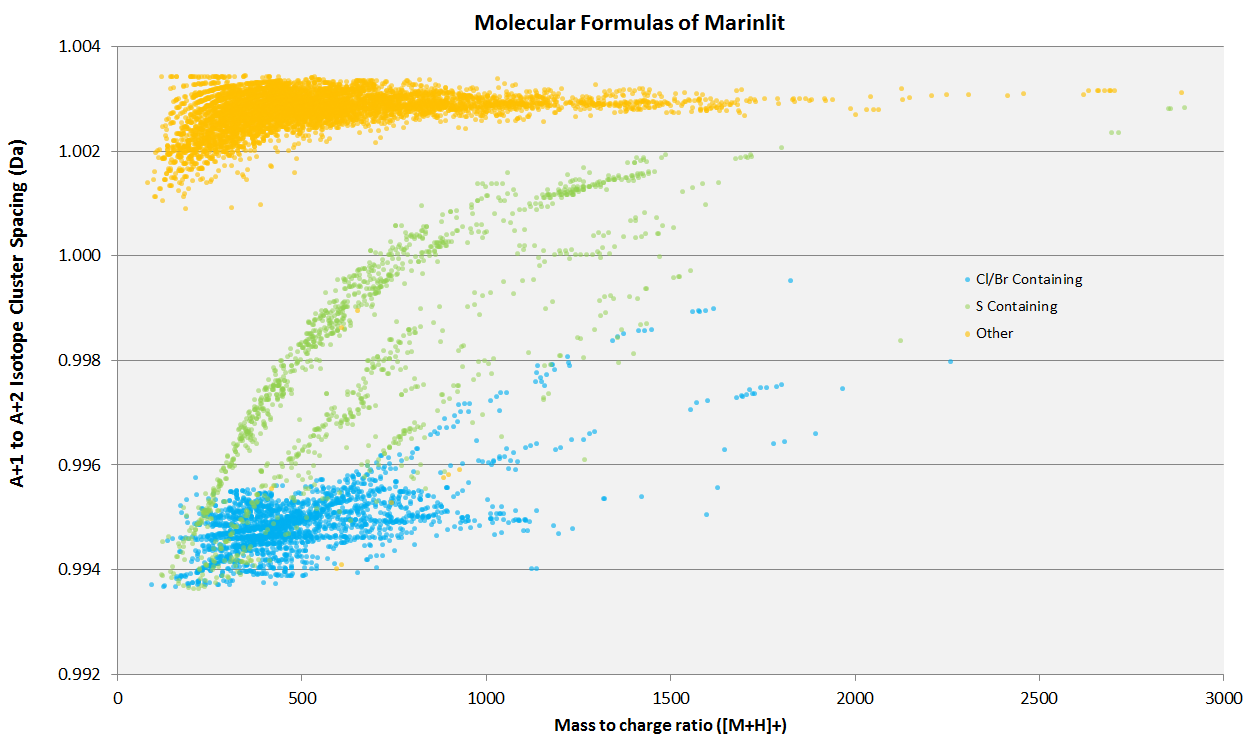
The A+1 to A+2 isotope cluster spacing of the simulated [M+H]+ isotope patterns of the molecular formulas of the Marinlit natural products database. Blue points are molecular formulas which contain either chlorine or bromine. Green points are molecular formulas which contain sulphur. Yellow points are molecular formulas which do not contain chlorine, bromine or sulphur.
Contact
Aaron John Christian Andersen Metabolomics Core Manager Phone: +45 45256138 ajca@dtu.dk